---
title: "Example 03: Experimenting with geoms"
---
```{r}
library(tidyverse)
library(gapminder)
library(palmerpenguins)
```
# Penguin histograms
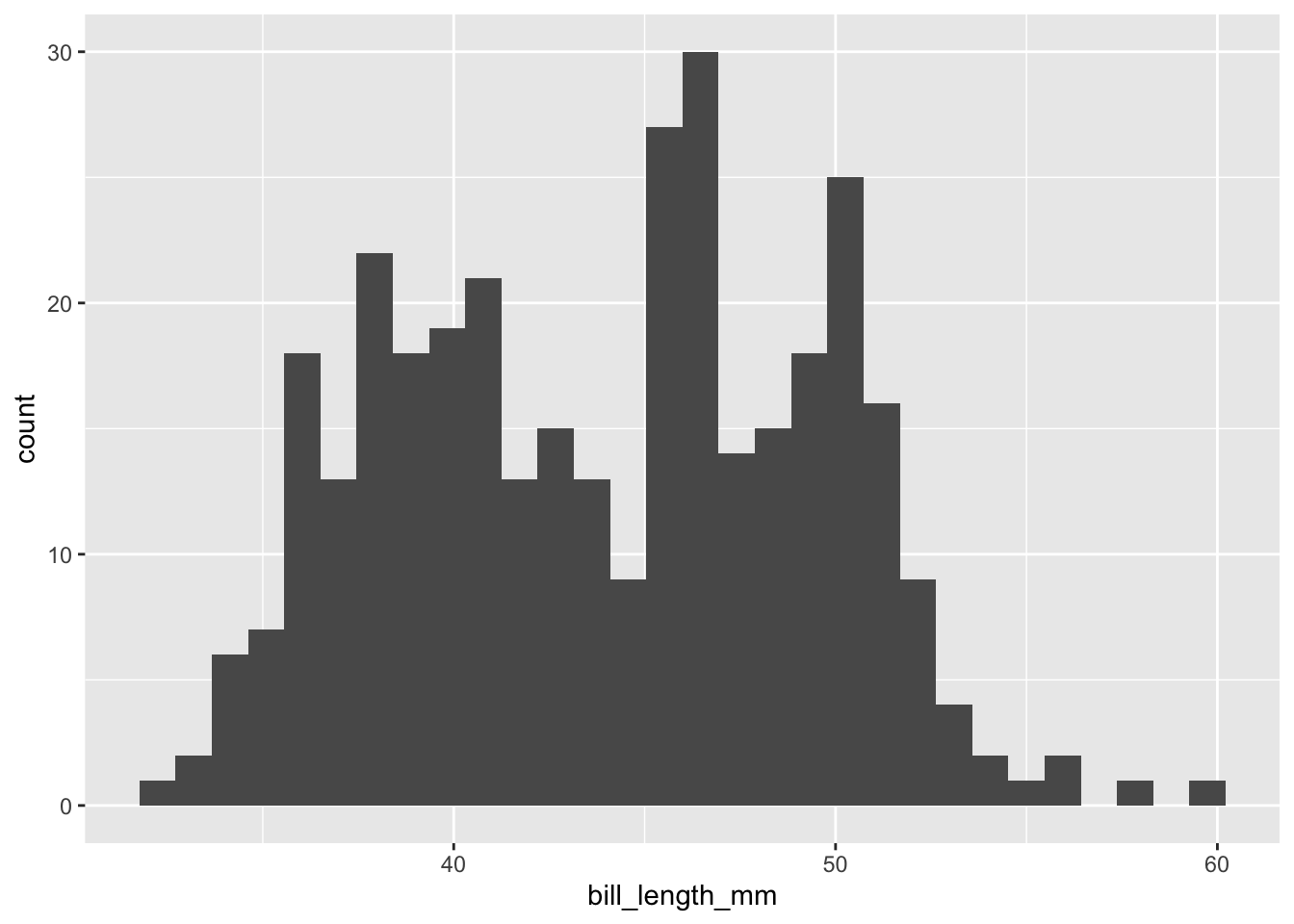
```{r}
penguins |>
ggplot(mapping = aes(bill_length_mm)) +
geom_histogram()
```
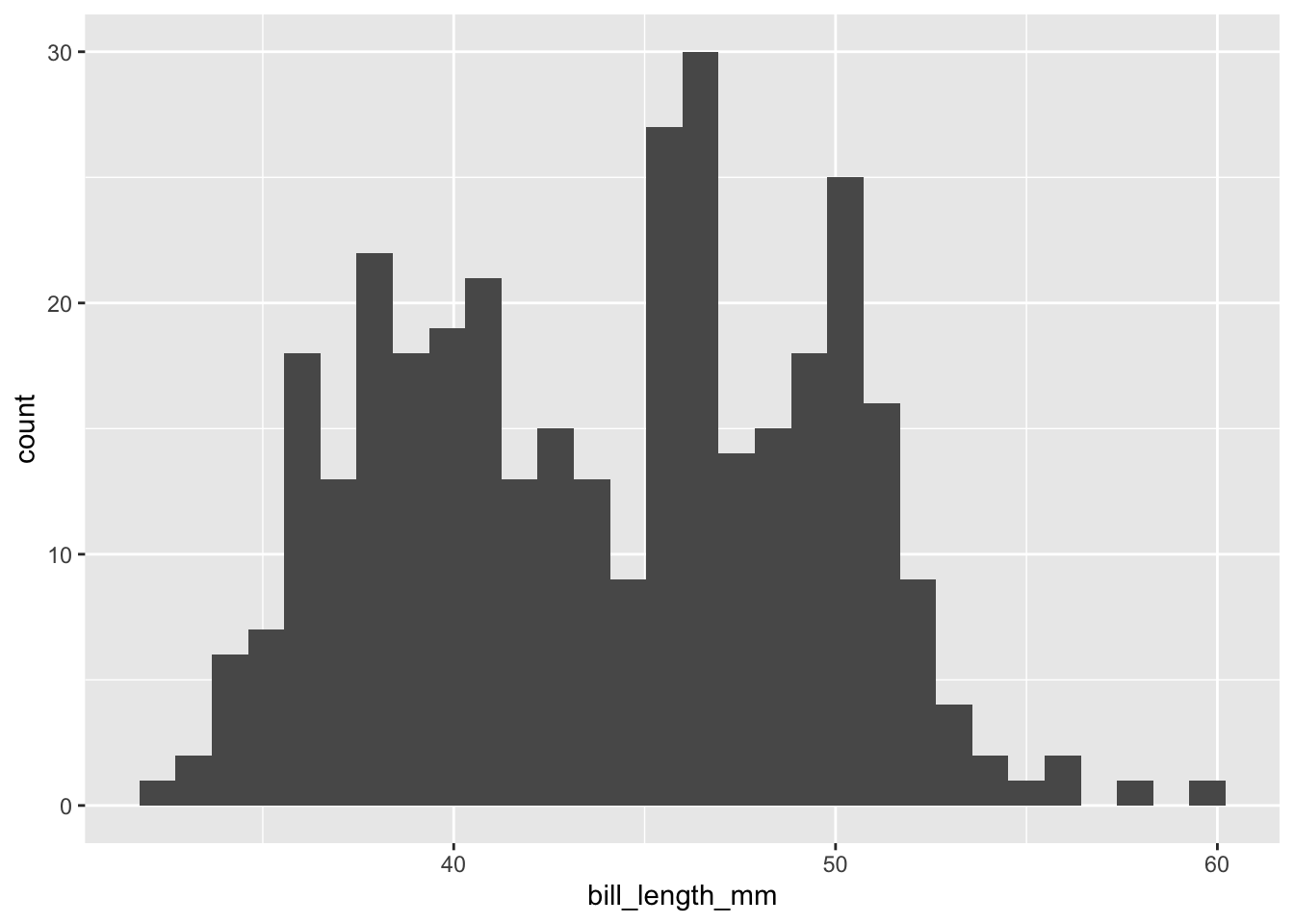
```{r}
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm)) +
geom_histogram()
```
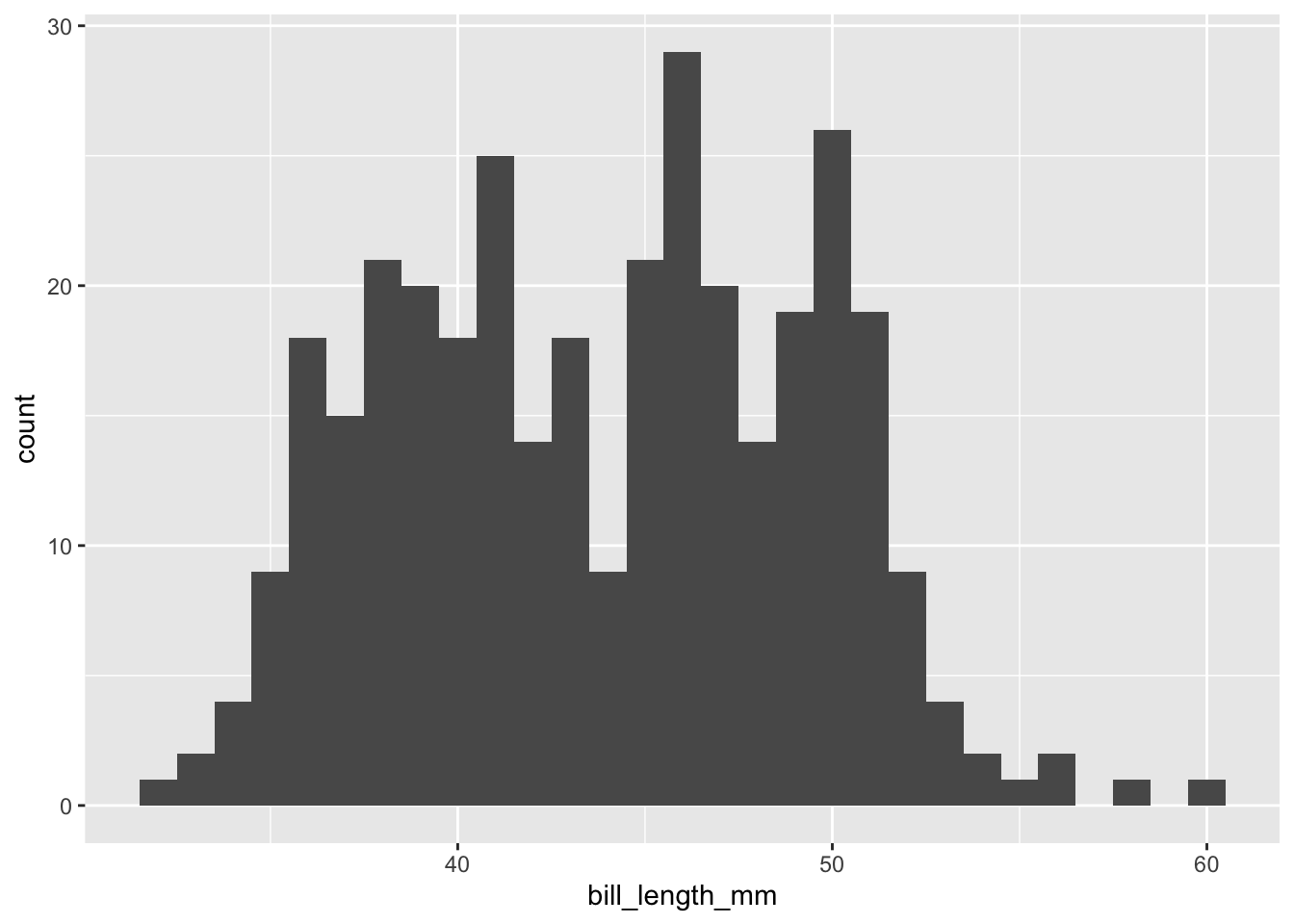
```{r}
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm)) +
geom_histogram(binwidth = 1)
```
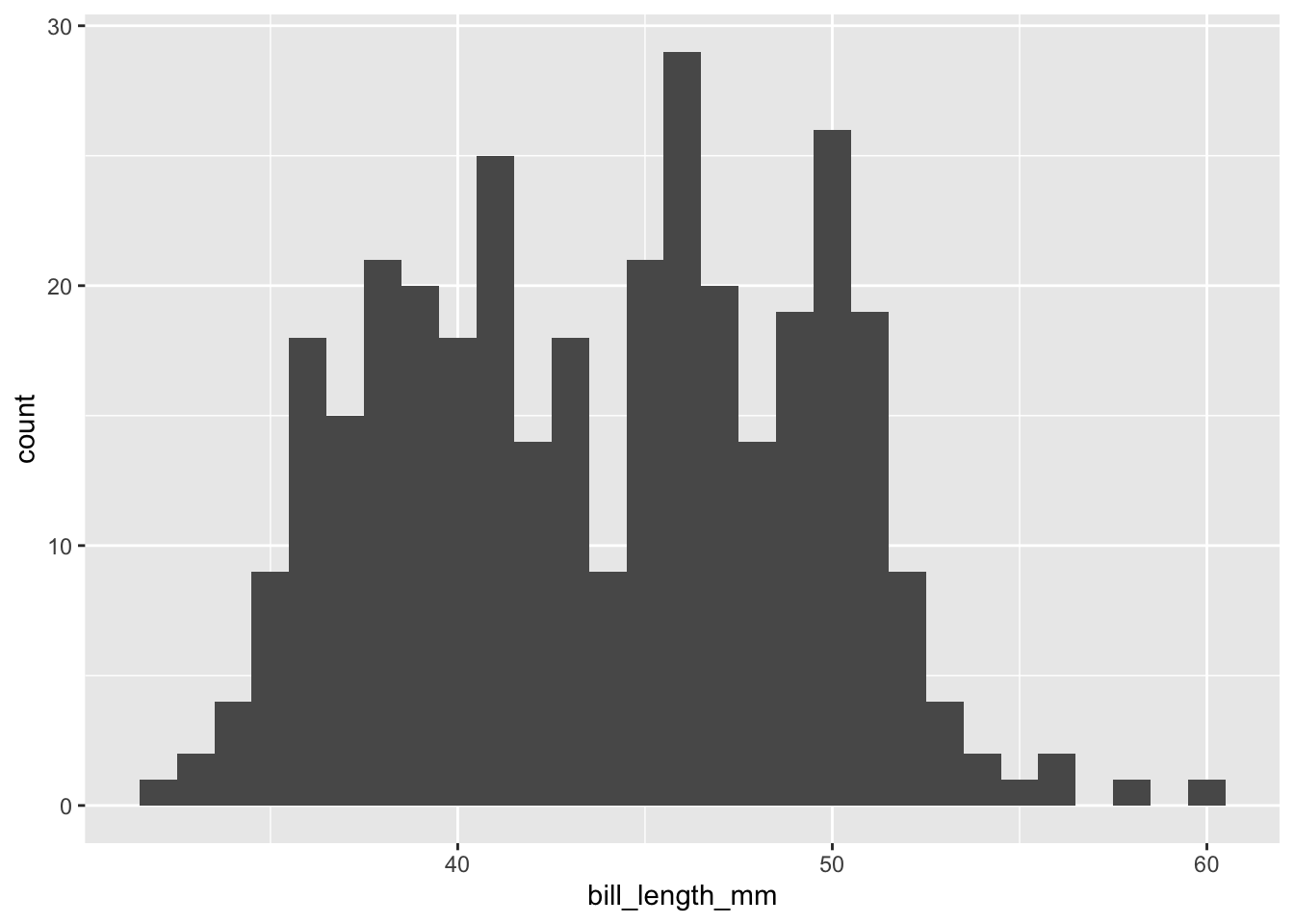
```{r}
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm)) +
geom_histogram(binwidth = 1)
```
```{r}
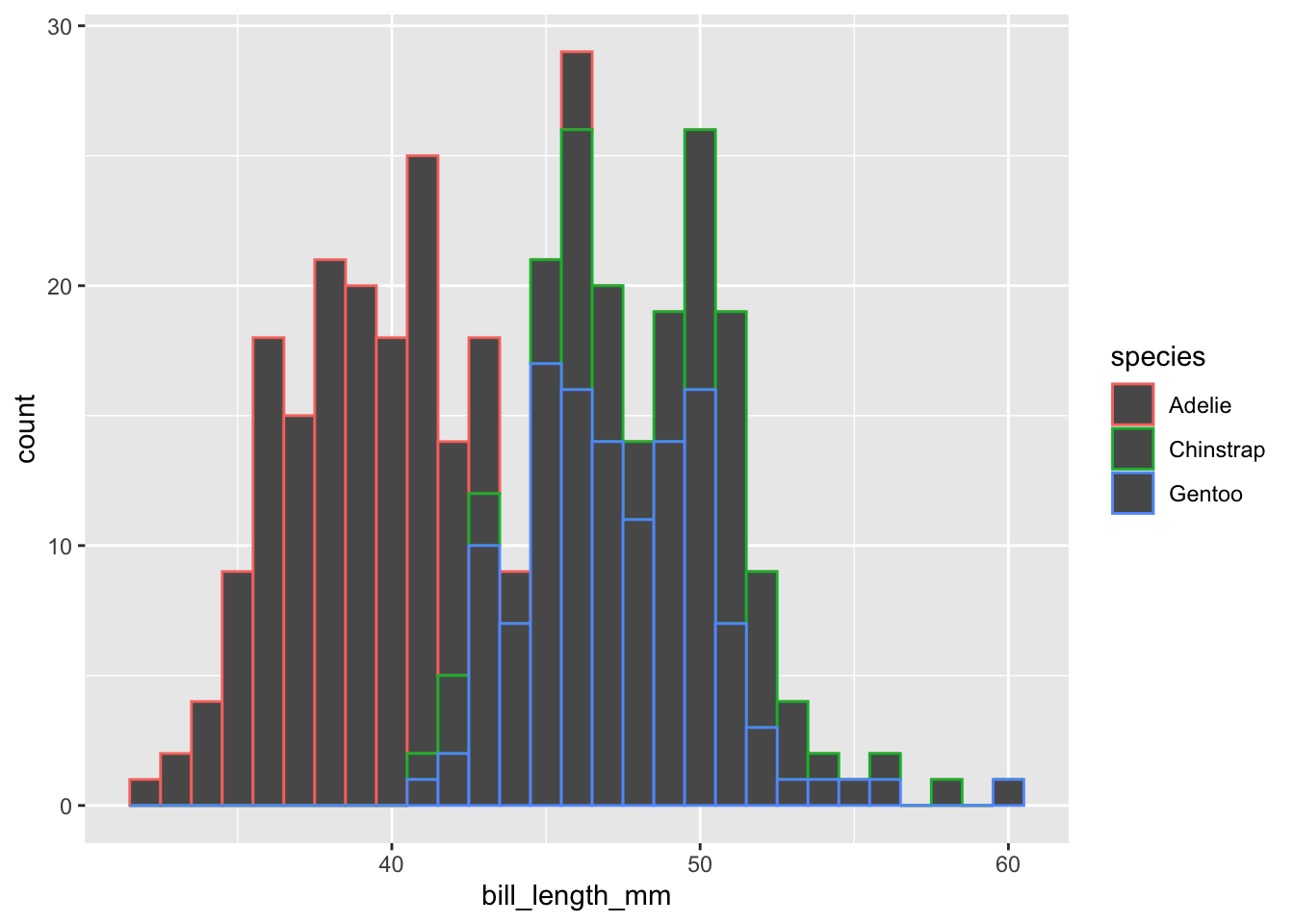
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm,
color = species)) +
geom_histogram(binwidth = 1)
```
```{r}
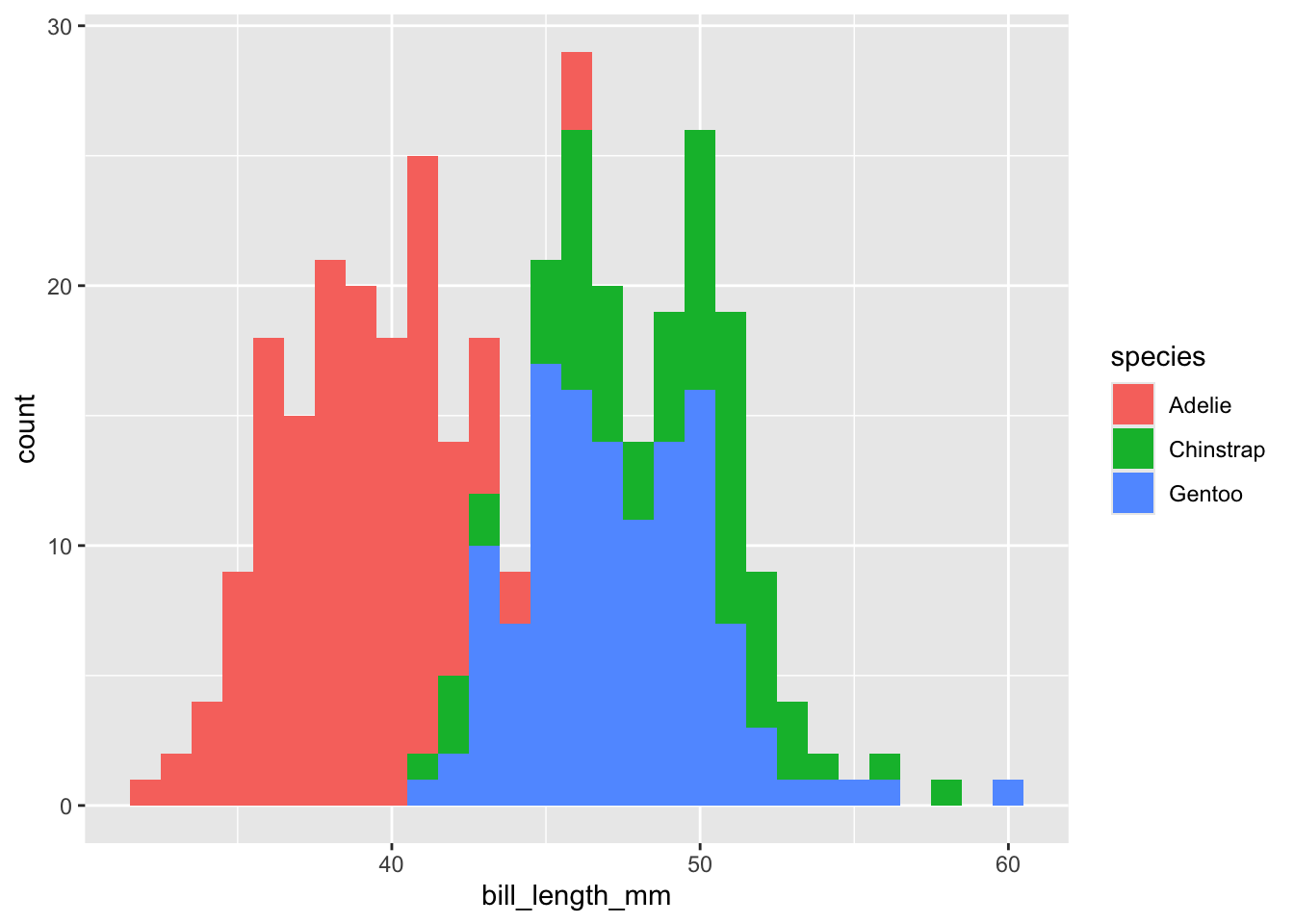
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm,
fill = species)) +
geom_histogram(binwidth = 1)
```
```{r}
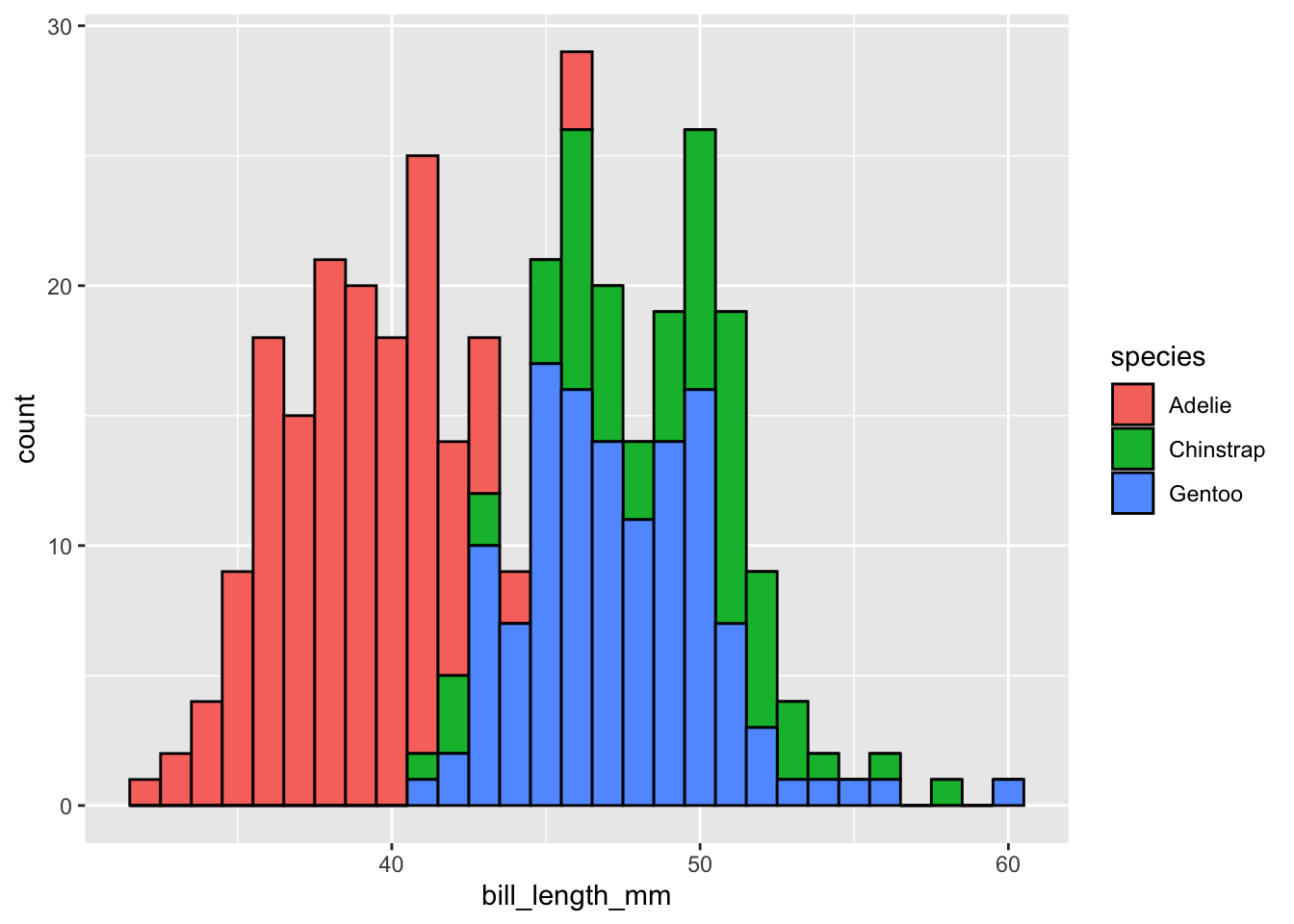
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm,
fill = species)) +
geom_histogram(binwidth = 1, color = "black")
```
```{r}
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm,
fill = species)) +
geom_histogram(binwidth = 1, color = "black") +
facet_wrap(~ species, ncol = 1)
```
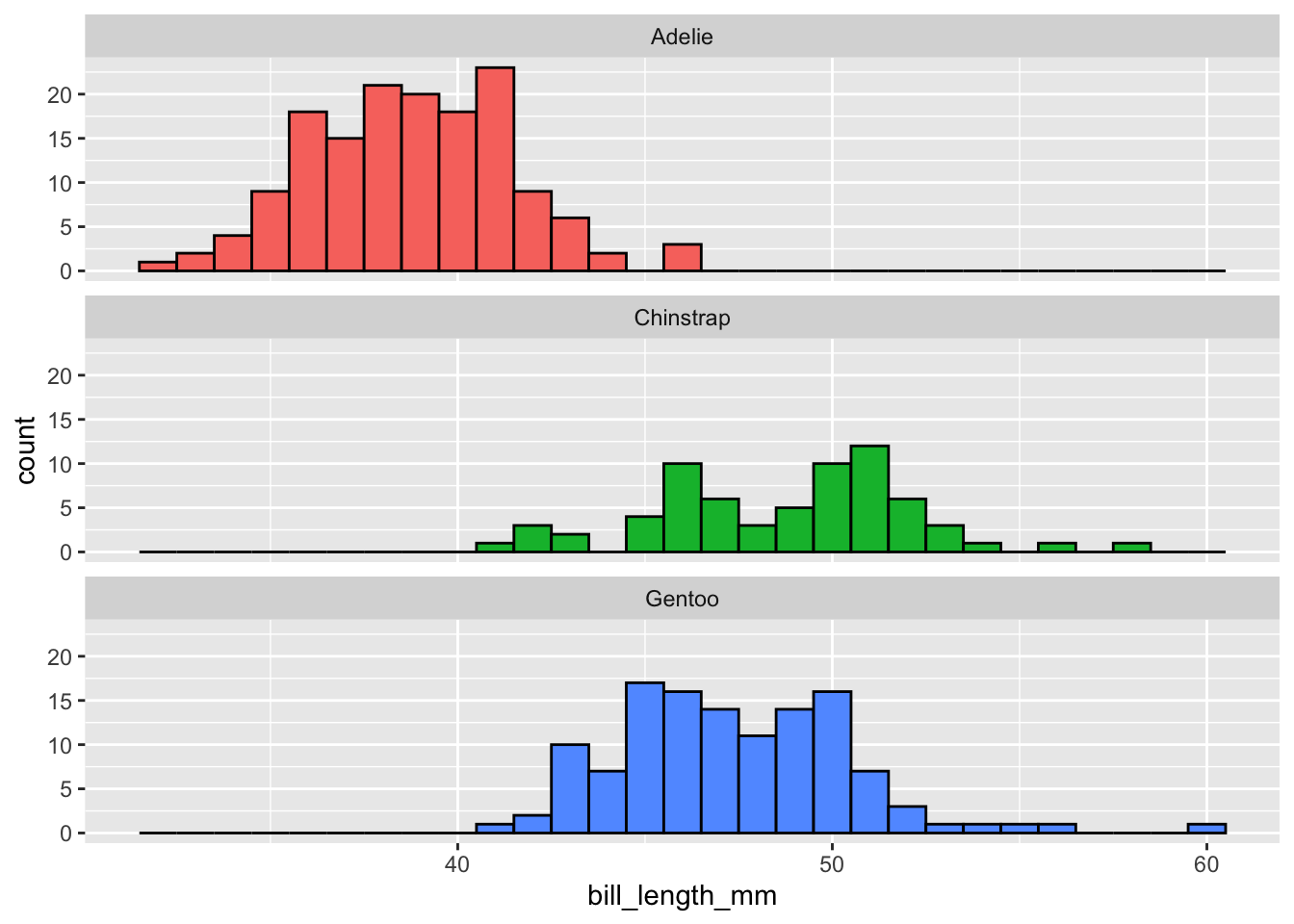
```{r}
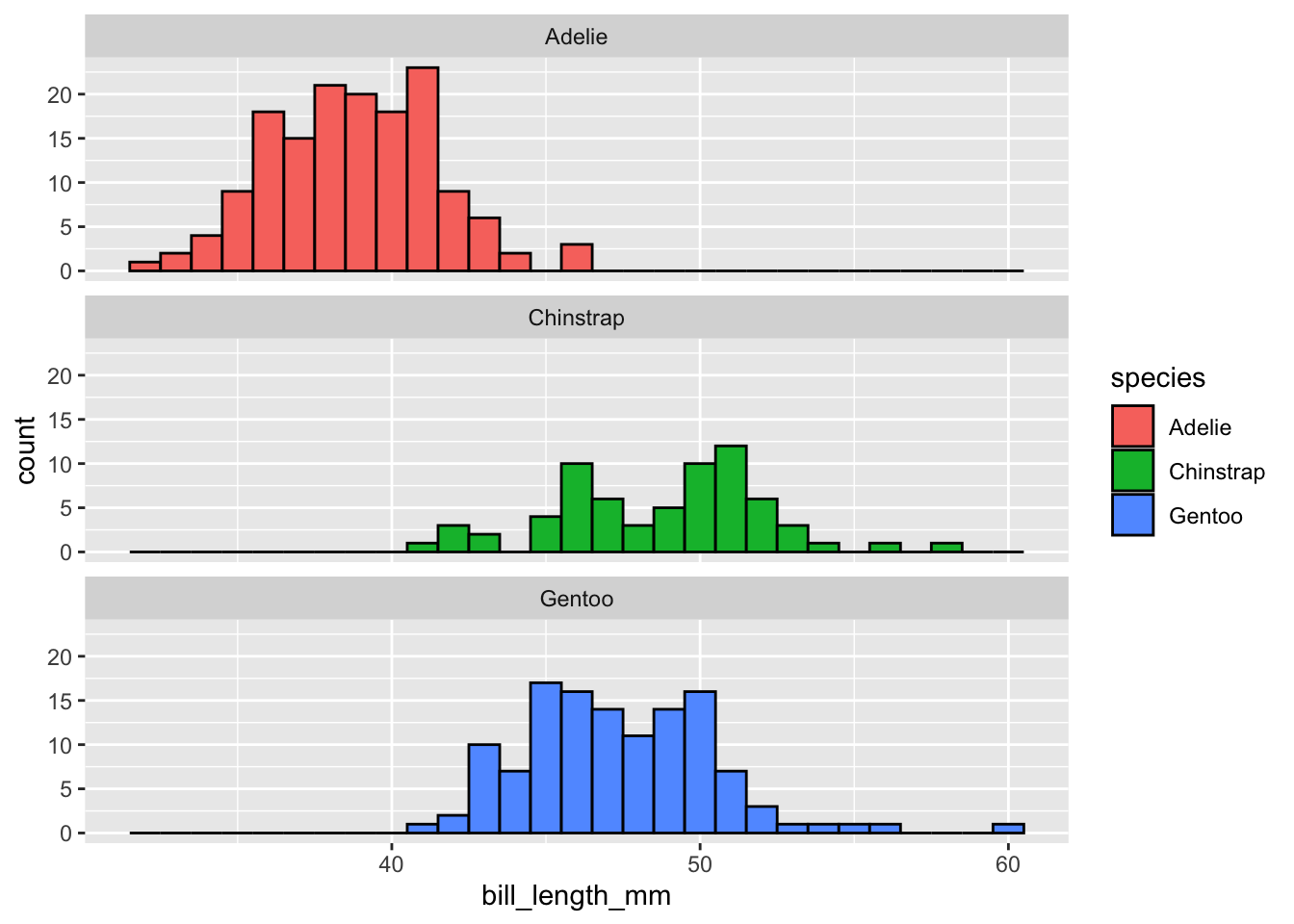
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm,
fill = species)) +
geom_histogram(binwidth = 1, color = "black") +
facet_wrap(~ species, ncol = 1) +
guides(fill = "none")
```
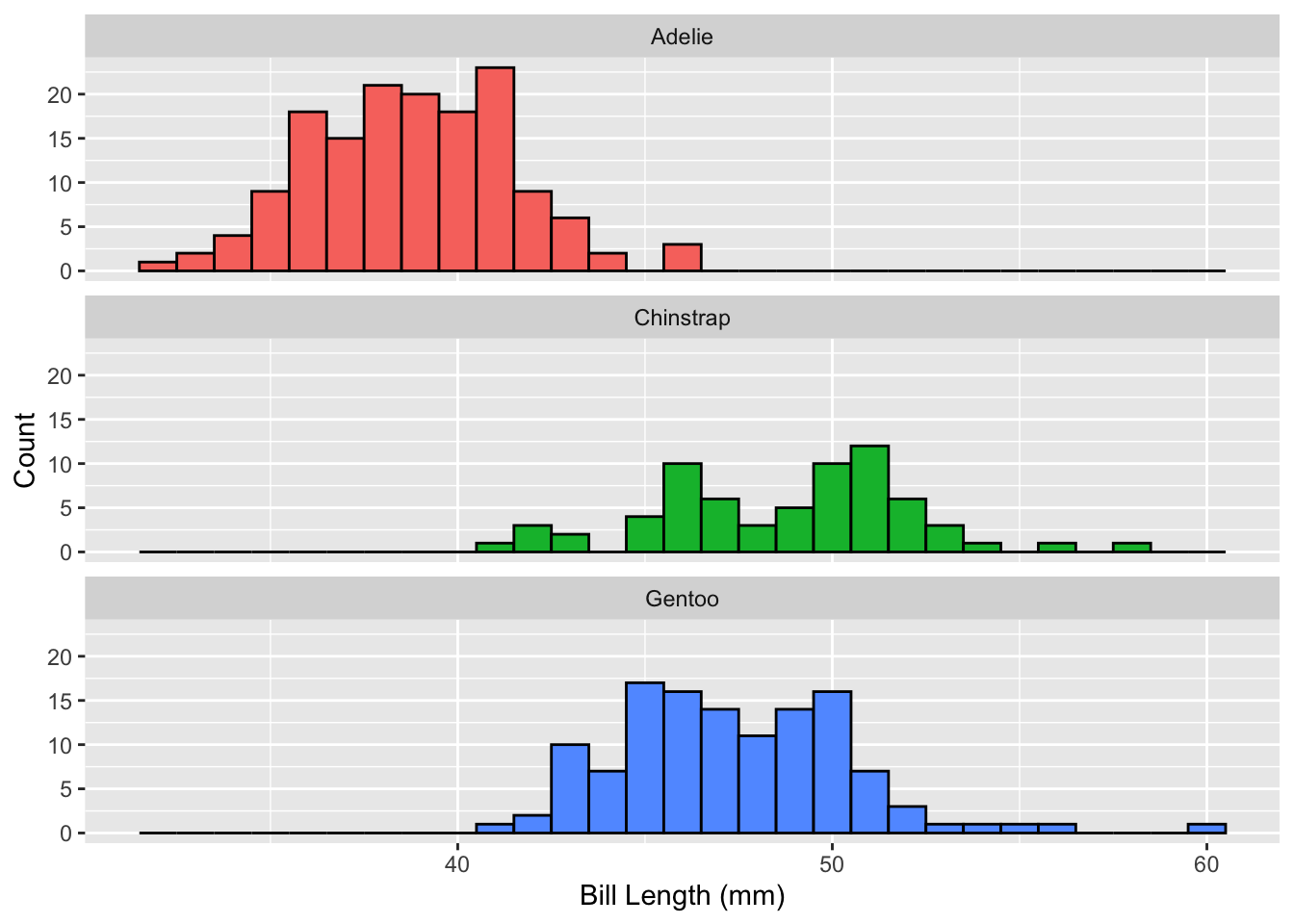
```{r}
penguins |>
drop_na(bill_length_mm) |>
ggplot(mapping = aes(x = bill_length_mm,
fill = species)) +
geom_histogram(binwidth = 1, color = "black") +
facet_wrap(~ species, ncol = 1) +
guides(fill = "none") +
labs(x = "Bill Length (mm)",
y = "Count")
```